I am a PhD student in Bioengineering at University of Pennsylvania, advised by Prof. Cesar de la Fuente. Previously, I received my bachelor's degree in Artificial Intelligence from Huazhong University of Science and Technology. I am fortunate to work under the supervision of Prof. Jianyang Zeng at Westlake University, Prof. Xiaohui Xie in University of California, Irvine and Prof. Dongrui Wu in Huazhong University of Science and Technology.
In the era of large language models, my research objective is to build models that can interact with and truly understand the real world—both the macroscopic world of physical objects and the microscopic 3D molecular world—so as to develop a knowledgeable, reliable AI biochemical expert capable of autonomous scientific discovery. To this end, my previous work has centered on:
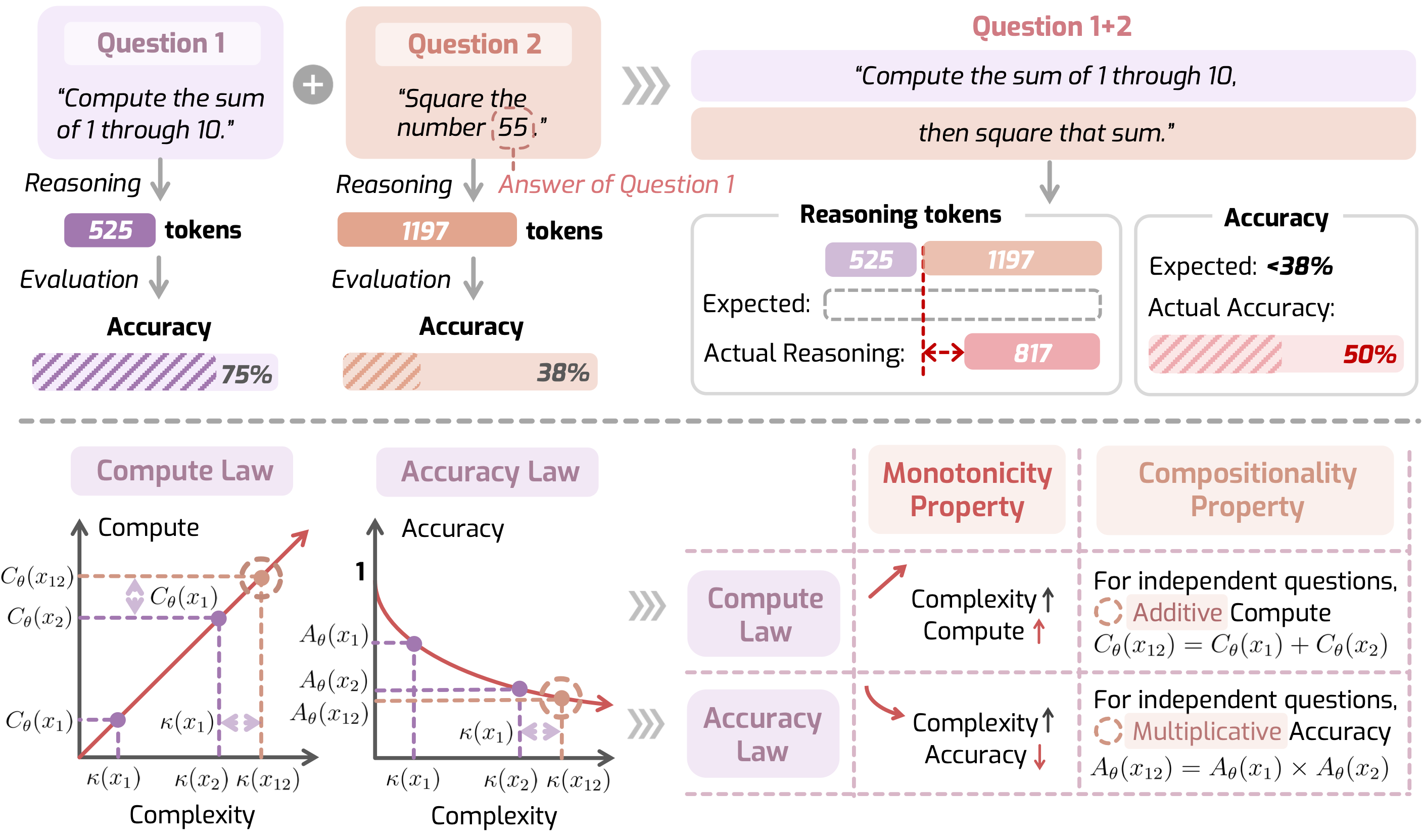
- Language model reasoning: how task or problem difficulty systematically affects the required reasoning depth/trajectory of LLMs and how this in turn influences accuracy.
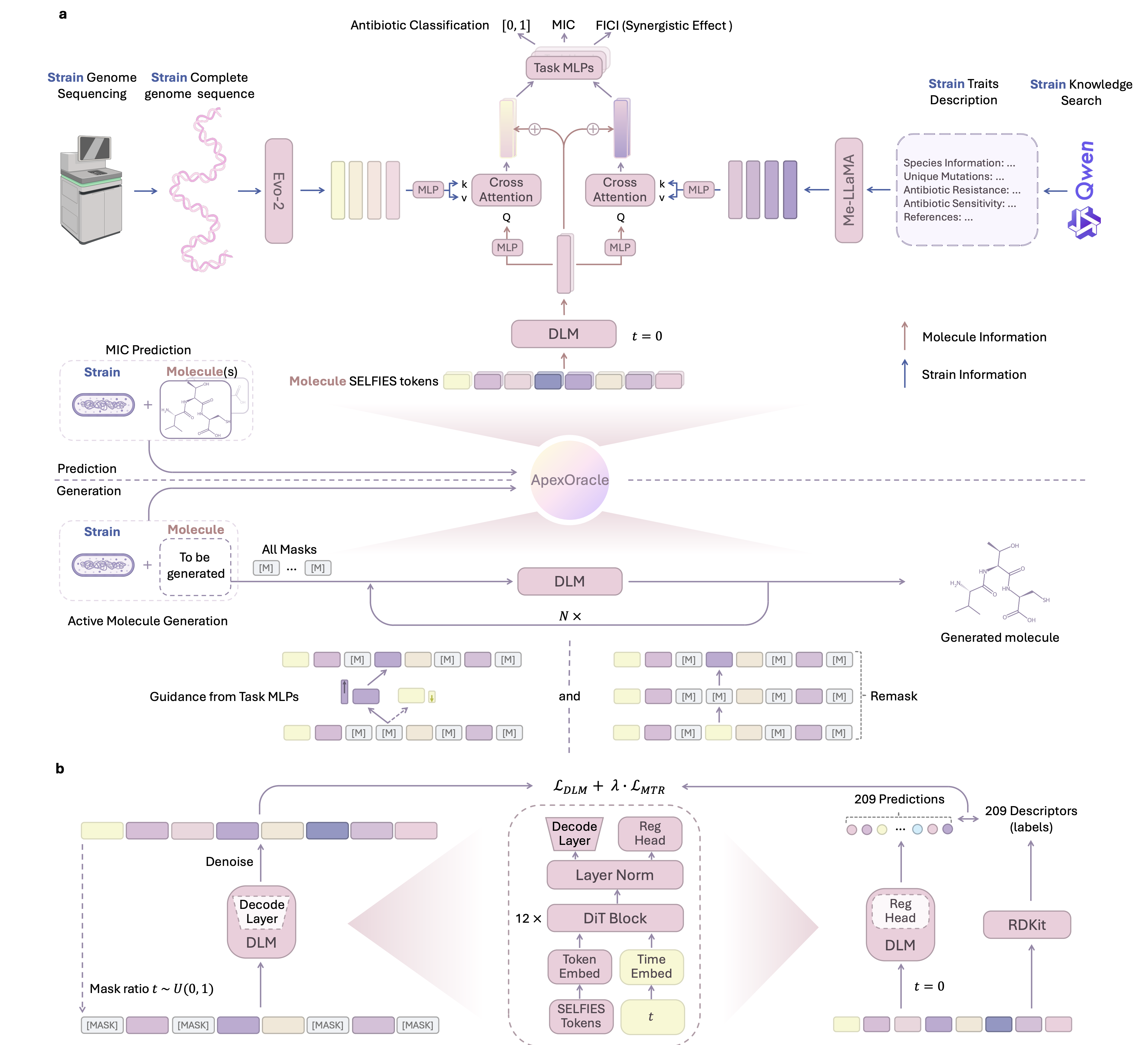
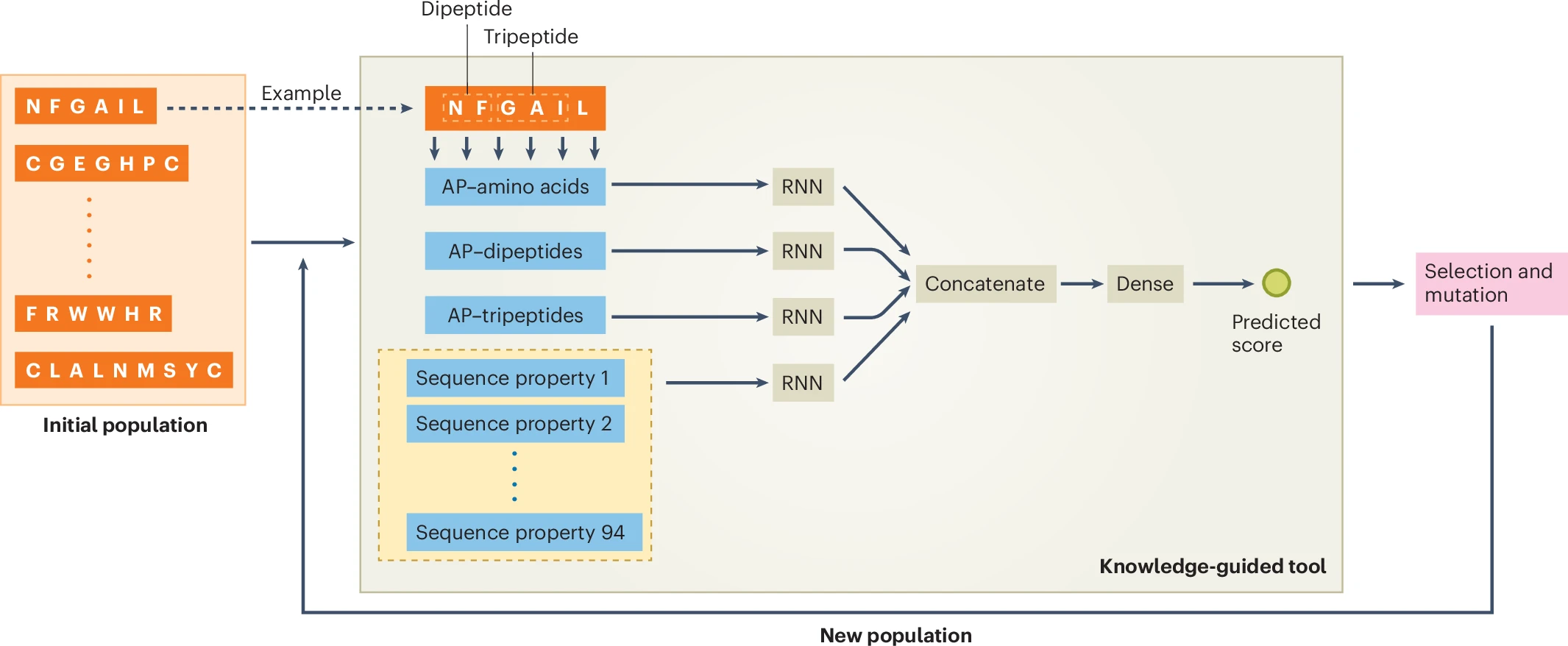
- Molecular understanding and generation: the advantages of discrete diffusion–style language models for molecular representation and design, featuring applications to antimicrobial peptides design.
- Medical image analysis: few-shot, high-precision segmentation of pathological organs. Going forward, this focus should be switched to vision-language models for cell-level perturbation–response understanding, coupled with automated, grounded natural-language reasoning outputs to close the loop from observation to scientific hypothesis.